HARMONY 2026
Conference Date
2 - 5 February 2026Conference Location
University of Auckland, Auckland, New ZealandImportant Dates
Important Links

The “Computational Modeling in Biology” Network (COMBINE) is an initiative to coordinate the development of the various community standards and formats in systems biology, synthetic biology and related fields. HARMONY is a codefest-type meeting, with a focus on development of the standards, interoperability and infrastructure. There are generally not many general discussions or oral presentations during HARMONY; instead, the time is devoted to allowing hands-on hacking and interaction between people focused on practical development of software and standards.
HARMONY 2026 will be held at the University of Auckland.
Local organizers are David Nickerson, Weiwei Ai, Jarrah Dowrick, Alan Garny, and Hugh Sorby at the Auckland Bioengineering Institute.
Auckland Human Digital Twin Meetings 2026
From 2–12 Feb 2026, the Auckland Bioengineering Institute hosts three workshops in Auckland advancing digital twin modelling, interoperability standards, and clinical translation.
On this page you can find conference accommodation rates, venue and transport information for getting around Auckland, details on the other workshops happening during the same period, information about Waitangi Day — New Zealand’s national holiday, which falls during the meetings; as well as an optional Waiheke Island excursion and plenty of things to do and places to dine around the city.
Workshop Location
HARMONY 2026 will be held at the University of Auckland. It will be co-located with the 2026 Cardiopulmonary Physiome Workshop.
The workshop will take place at:
The Faculty of Engineering (Building 401) Atrium (Room 418) 20 Symonds Street, Auckland Central.
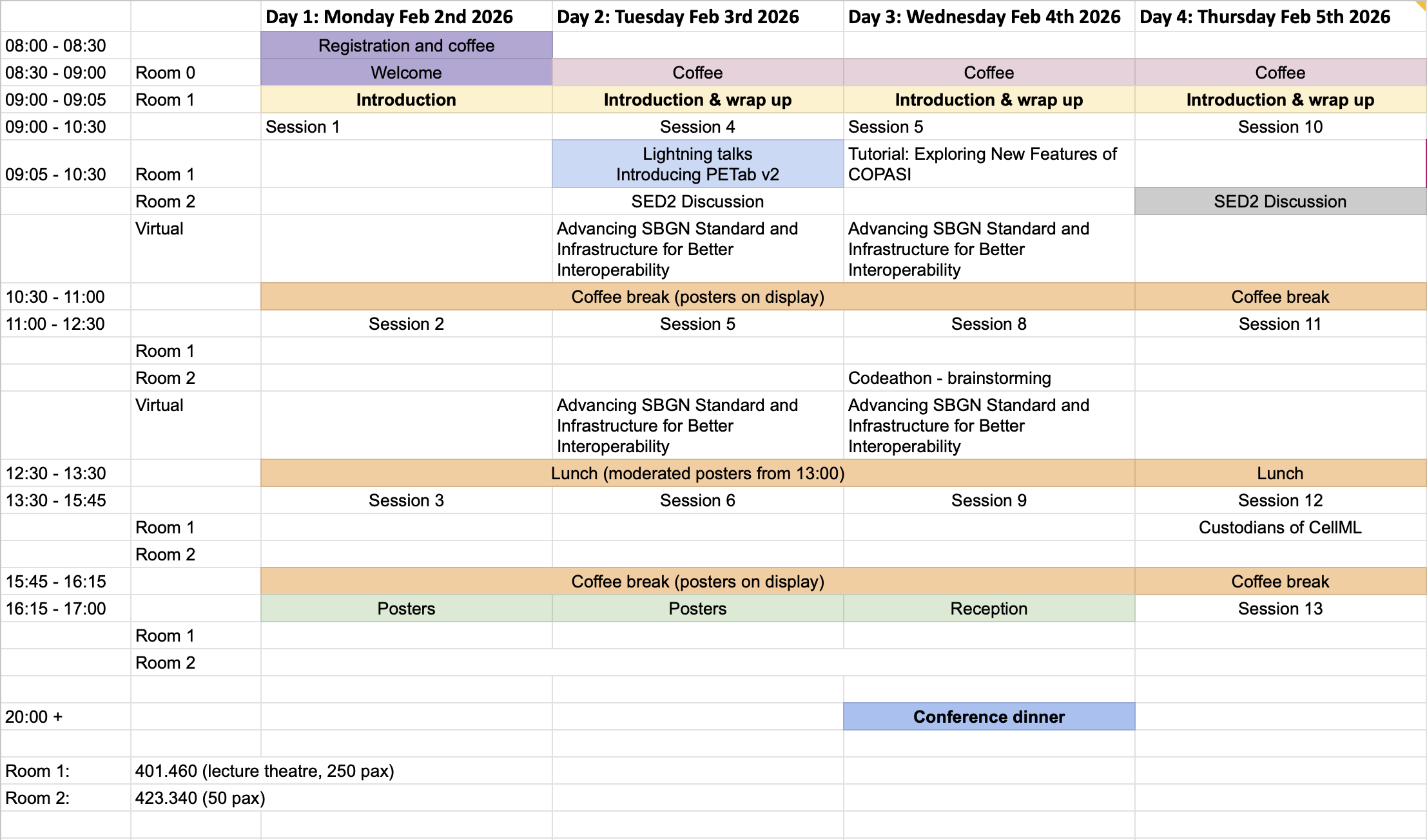
Current Schedule
Arrival and Transportation
Getting from Auckland Airport (AKL) to the central city (also known as the ‘CBD’) is straightforward. Your main options depend on your budget, time, and preference for convenience.
Quick Comparison Table
| Method | Approx. Cost (1 Adult) | Approx. Time | Best For |
|---|---|---|---|
| Public Transport | ~$6.50 | 50 - 60 mins | Budget |
| Express Bus (SkyDrive) | ~$18 | 35 - 50 mins | Simplicity & Value |
| Ride-Share (Uber) | ~$40 - $65 | 25 - 45 mins | On-Demand Convenience |
| Taxi (Fixed Fare) | ~$62 - $99 | 20 - 40 mins | Speed & Ease |
| Shared Shuttle | ~$25 - $40 | 45 - 75 mins | Door-to-Door Value |
Note: Travel times can be heavily affected by traffic, especially during morning (7:00 AM - 9:30 AM) and evening (4:00 PM - 6:30 PM) peak hours.
Visitor Tip: Always ask for a fixed fare or flat rate to the city before you get in. Metered fares can be much more expensive (up to $90-$100+ in bad traffic).
Further information on these transport options is available in the following places:
Accommodation
Accommodation has been combined with the Human Digital Twin meetings and information on accommodation can be found there.
Human Digital Twin meetings accommodation information
Meals
Meals during the day for Harmony 2026 will be provided in coordination with the other digital twins meetings. For evening meals there is a great selection of a wide variety of restaurants a short walk or short taxi/Uber from the conference location.
Power outlet
Type I plugs are used in New Zealand, featuring three flat pins arranged in a triangular pattern. They operate on a voltage of 230V and are designed to ensure safety with an earth pin for grounding.
Plugs used in New Zealand (Wikipedia).

Attendees
The following are the registered attendees for HARMONY 2026, as of 2026-01-29 01:28 UTC.
| Name | Affiliation | Attendance | Interests |
|---|---|---|---|
| Hugh Sorby | Auckland Bioengineering Institute | In person | CellML, SED-ML |
| Falk Schreiber | University of Konstanz | In person | SBGN, SBML, SBOL and SBOL Visual |
| Alan Garny | University of Auckland | In person | CellML, COMBINE Archive, Multicellular Modelling, OMEX Metadata, SED-ML |
| Weiwei Ai | Auckland Bioengineering Institute, University Of Auckland | In person | CellML, SED-ML, PE-TAB |
| Xiaoming Hu | Bioquant, University Heidelberg | In person | SBML, Data Management, Ro-Crate |
| Lucian Smith | University of Washington | In person | COMBINE Archive, Multicellular Modelling, OMEX Metadata, SBML, SED-ML, FROG |
| Chris Myers | University of Colorado Boulder | In person | COMBINE Archive, SBML, SBOL and SBOL Visual, SED-ML |
| Frank T. Bergmann | BioQUANT, Heidelberg University | In person | COMBINE Archive, SBGN, SBML, SED-ML, PE-TAB, FROG |
| David Nickerson | Auckland Bioengineering Institute, University of Auckland | In person | CellML, COMBINE Archive, Multicellular Modelling, OMEX Metadata, SED-ML, PE-TAB, repositories; model publishing |
| Eric Young | WPI | In person | SBOL and SBOL Visual |
| Triana Karnadipa | ABI | Remotely | COMBINE Archive, NeuroML, OMEX Metadata |
| Pedro T. Monteiro | IST - University of Lisbon | Remotely | Multicellular Modelling, SBML, SED-ML, FROG |
| MORETTI Sébastien | SIB Swiss Institute of Bioinformatics | Remotely | BioPAX, SBGN, SBML |
| Ayesha Abubakr | St. John’s Kilmarnock School | Remotely | BioPAX, CellML, Multicellular Modelling, NeuroML, SBML, SBOL and SBOL Visual, SED-ML |
| Gonzalo Vidal | University of Colorado Boulder | Remotely | SBOL and SBOL Visual |
| Hoda Rahimi | The University of Auckland | Remotely | BioPAX, CellML, COMBINE Archive, Multicellular Modelling, OMEX Metadata, SBGN, SBML, SED-ML, PE-TAB |
| Litian Su | Auckland Bioengineering Institute | Remotely | CellML, SBML |
| Maria Chiara Langella | University of Naples Federico II | Remotely | BioPAX, CellML, COMBINE Archive, Multicellular Modelling, SBML, SBOL and SBOL Visual, PE-TAB |
| Leo Willyanto Santoso | Auckland Bioengineering Institute, The University of Auckland | Remotely | CellML |
| Koray Atalag | Auckland Bioengineering Institute | Remotely | health data interoperability (FHIR, openEHR, SNOMED, LOINC, ICD etc.) |
| Lutz Brusch | TU Dresden, Germany | Remotely | Multicellular Modelling, SBML, PE-TAB, OpenVT, MultiCellML, MorpheusML |
| Jarrah Dowrick | Auckland Bioengineering Institute | In person | CellML, COMBINE Archive, OMEX Metadata, SBGN, SED-ML, FieldML |
| Goksel Misirli | Keele University | Remotely | Multicellular Modelling, OMEX Metadata, SBGN, SBML, SBOL and SBOL Visual |
| University at Buffalo | Remotely | ||
| Marc Molla Garcia | UPF | In person | BioPAX, CellML, COMBINE Archive, Multicellular Modelling, NeuroML, OMEX Metadata, SBGN, SBML, SBOL and SBOL Visual, SED-ML, PE-TAB, FROG |
| Matthias König | Humboldt-Universität zu Berlin, Faculty of Life Sciences, Department of Biology, Institute of Theoretical Biology, Systems Medicine of the Liver; University of Stuttgart, Institute of Structural Mechanics and Dynamics in Aerospace Engineering | Remotely | CellML, COMBINE Archive, Multicellular Modelling, OMEX Metadata, SBGN, SBML, SED-ML, PE-TAB, FROG |
| Augustin Luna | National Library of Medicine | Remotely | BioPAX, SBGN |
| Hasan Balci | National Library of Medicine, NIH | Remotely | SBGN, SBML |
| Fengkai Zhang | NIH, US | Remotely | COMBINE Archive, Multicellular Modelling, OMEX Metadata, SBGN, SBOL and SBOL Visual, SED-ML, PE-TAB |
| Te Chen | Technical University of Denmark | Remotely | SBML, SED-ML |
| Viola Rintoul | UW Madison | Remotely | CellML, COMBINE Archive, Multicellular Modelling, SBOL and SBOL Visual, SED-ML |
| Rahuman Sheriff | EMBL-EBI | Remotely | CellML, COMBINE Archive, Multicellular Modelling, OMEX Metadata, SBGN, SBML, SED-ML, PE-TAB, FROG |
Registration
There is a NZ$640.00 registration fee for in-person attendess, please use the links available in the registration form to pay via Eventbrite and sign up for any add-on activities. For virtual attendess, there is no fee but you do need to register. Also, please register using the link provided as soon as possible. This will help us plan the schedule and match your interests to the timing of the breakouts, etc. Note, only registered attendees will be sent information related to the meeting.Call for Breakout Sessions and Tutorials
All attendees can suggest breakout sessions for hacking and/or detailed discussions of certain aspects of one or several of the COMBINE standard(s), metadata and semantic annotations (format-specific or overarching), application and implementations of the COMBINE standards, or any other topic relevant for the COMBINE community. The topics for those breakout sessions, and the time slots which would suit their communities can be submitted via the link above. Note, breakout session organisers will be responsible for creating and hosting their own online sessions, if required.Call for Lightning Talks and Posters
Requests for a lightning talk (5 min max.) and/or poster can be submitted via the link above. Please make multiple submissions if you want to submit abstracts on different topics. Talks will take place during the community session and posters will be displayed throughout the meeting.Topics of Interest
- Data exchange, pipelines and model standards for systems and synthetic biology
- Visualisation and graphical notation standards for systems and synthetic biology
- Standards for sharing and analysing biological pathway data
- Standards for computational biological models and modelling support
- Metadata description and model annotation in COMBINE standard formats
- Implementation of COMBINE standards in tools, databases and other resources
- Integrated model and data management for systems and synthetic biology
- Standardization of Artificial Intelligence approaches in biological modelling
- Emerging standardization needs and multicellular modeling
- Community aspects of COMBINE